[1] 6Efficient R Programming
Working with Many Columns, Functions, and Models
CSSCR & Political Science, UW
Introduction
Rule of thumb: “consider writing a function whenever you’ve copied and pasted a block of code more than twice”
What is in a function?
- Input → Output
In R, you can write a named function
Introduction
Benefits of functional programming
Reduce redundancy & improve readability
- Less copy-and-paste and mistakes; more reusable
Encourage modular thinking
- Break complex workflows into small, testable functions
Enhance scalability
- Apply across many observations, columns or datasets
tidyverse pipelines friendly
- Integrate with
mutate(),across(),map()
- Integrate with
Operations in R
- Step back and think about basic operations in R
Operations in R
- What if I want to add 1 to each and every value?
- Why it works?
Operations in R
- It’s like having two vectors of the same length
- And adding two vectors together at once
Operations in R
Vectorized operation is extremely fast and efficient
Operations in R
- Let’s go to another extreme: for loop
Operations in R
It iterates through each and every value
- It’s generally slower and involves more memory & computational overhead
Operations in R
Vectorized operation
- “Do this operation on the whole vector at once!”
For loop
- “Do this, then this, then … – repeat manually for each element!”
Functional programming
- “Define a function and apply it automatically to each element or column!”
Functions and map()
Functions and map()
Functions and map()
Why uses map()?
- B/c many operations don’t work with vectors
- It doesn’t work on vectors
Functions and map()
- But
map()works
Functions and map()
- A more realistic example: a list of vectors
$`Group A`
[1] -0.3901175 -0.7516687 1.1894436 0.2955950 -0.5227733
$`Group B`
[1] 0.8254738 0.7300041 -0.3148819 -0.9441314 -0.2647824
$`Group C`
[1] 0.2754437 -1.1706525 -0.5063522 1.1297897 -1.9233868[1] "list"Functions and map()
- A more realistic example: a list of vectors
Warning in mean.default(some_data): argument is not numeric or logical:
returning NA[1] NA- Why error?
Functions and map()
- A more realistic example: a list of vectors
Functions and map()
- Why if you need more flexibility from functions?
Functions and map()
- Define a new function that takes care of NAs
Functions and map()
- Or, you can directly write an anonymous function
Functions and map()
- Or, using shorthand
~(lambda) and.x(placeholder)
Wrangling multiple columns
- Iris example
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 140 more rowsWrangling multiple columns
- Round up one column
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 3.5 1.4 0.2 setosa
2 5 3 1.4 0.2 setosa
3 5 3.2 1.3 0.2 setosa
4 5 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5 3.9 1.7 0.4 setosa
7 5 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4 2.9 1.4 0.2 setosa
10 5 3.1 1.5 0.1 setosa
# ℹ 140 more rowsWrangling multiple columns
- How about multiple columns?
iris %>%
mutate(Sepal.Length = round(Sepal.Length),
Sepal.Width = round(Sepal.Width),
Petal.Length = round(Petal.Length),
Petal.Width = round(Petal.Width))# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 4 1 0 setosa
2 5 3 1 0 setosa
3 5 3 1 0 setosa
4 5 3 2 0 setosa
5 5 4 1 0 setosa
6 5 4 2 0 setosa
7 5 3 1 0 setosa
8 5 3 2 0 setosa
9 4 3 1 0 setosa
10 5 3 2 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- Using
across()with function
iris %>%
mutate(across(.cols = c(Sepal.Length, Sepal.Width, Petal.Length, Petal.Width),
.fns = round))# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 4 1 0 setosa
2 5 3 1 0 setosa
3 5 3 1 0 setosa
4 5 3 2 0 setosa
5 5 4 1 0 setosa
6 5 4 2 0 setosa
7 5 3 1 0 setosa
8 5 3 2 0 setosa
9 4 3 1 0 setosa
10 5 3 2 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- For simplicity
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 4 1 0 setosa
2 5 3 1 0 setosa
3 5 3 1 0 setosa
4 5 3 2 0 setosa
5 5 4 1 0 setosa
6 5 4 2 0 setosa
7 5 3 1 0 setosa
8 5 3 2 0 setosa
9 4 3 1 0 setosa
10 5 3 2 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- Target specific columns
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 4 1.4 0.2 setosa
2 5 3 1.4 0.2 setosa
3 5 3 1.3 0.2 setosa
4 5 3 1.5 0.2 setosa
5 5 4 1.4 0.2 setosa
6 5 4 1.7 0.4 setosa
7 5 3 1.4 0.3 setosa
8 5 3 1.5 0.2 setosa
9 4 3 1.4 0.2 setosa
10 5 3 1.5 0.1 setosa
# ℹ 140 more rowsWrangling multiple columns
- Target specific columns
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 4 1.4 0 setosa
2 4.9 3 1.4 0 setosa
3 4.7 3 1.3 0 setosa
4 4.6 3 1.5 0 setosa
5 5 4 1.4 0 setosa
6 5.4 4 1.7 0 setosa
7 4.6 3 1.4 0 setosa
8 5 3 1.5 0 setosa
9 4.4 3 1.4 0 setosa
10 4.9 3 1.5 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- Target specific columns
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 4 1 0 setosa
2 5 3 1 0 setosa
3 5 3 1 0 setosa
4 5 3 2 0 setosa
5 5 4 1 0 setosa
6 5 4 2 0 setosa
7 5 3 1 0 setosa
8 5 3 2 0 setosa
9 4 3 1 0 setosa
10 5 3 2 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- Target specific columns based on another function
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5 4 1 0 setosa
2 5 3 1 0 setosa
3 5 3 1 0 setosa
4 5 3 2 0 setosa
5 5 4 1 0 setosa
6 5 4 2 0 setosa
7 5 3 1 0 setosa
8 5 3 2 0 setosa
9 4 3 1 0 setosa
10 5 3 2 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- Using anonymous function to further specify arguments
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 10 0 0 0 setosa
2 0 0 0 0 setosa
3 0 0 0 0 setosa
4 0 0 0 0 setosa
5 0 0 0 0 setosa
6 10 0 0 0 setosa
7 0 0 0 0 setosa
8 0 0 0 0 setosa
9 0 0 0 0 setosa
10 0 0 0 0 setosa
# ℹ 140 more rowsWrangling multiple columns
- A more realistic example: survey data
Rows: 215
Columns: 109
$ id <dbl> 3, 5, 6, 11, 13, 15, 21, 22, 23, 25, 26, 27, 28, 29, 30, 31,…
$ Q1_1 <dbl> 8, 35, 34, 20, 20, 36, 12, 11, 18, 24, 29, 21, 43, 10, 26, 2…
$ Q1_2 <dbl> 2.0, 12.0, 12.0, 9.0, 3.0, 20.0, 2.5, 0.5, 3.0, 8.0, 14.0, 5…
$ Q2 <ord> 2009, Before 2002, Before 2002, 2010, 2010, Before 2002, 200…
$ Q3_1 <fct> No, Yes, Yes, No, No, No, Yes, Yes, Yes, No, Yes, No, Yes, N…
$ Q3_2 <fct> No, No, Yes, No, No, Yes, No, Yes, Yes, No, Yes, Yes, Yes, Y…
$ Q3_3 <fct> No, No, No, No, No, No, No, Yes, Yes, No, Yes, Yes, No, No, …
$ Q3_4 <fct> No, No, No, No, No, No, No, Yes, Yes, Yes, Yes, No, No, No, …
$ Q3_5 <fct> No, No, No, No, No, No, No, Yes, Yes, Yes, Yes, No, No, Yes,…
$ Q3_6 <fct> No, No, Yes, No, No, No, Yes, No, Yes, Yes, Yes, No, Yes, No…
$ Q3_7 <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, No, No, No, No…
$ Q3_8 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q3_9 <fct> No, Yes, No, No, No, Yes, No, No, Yes, Yes, No, No, No, No, …
$ Q3_10 <fct> Yes, No, No, No, No, No, No, No, No, No, No, No, No, No, No,…
$ Q3_11 <fct> No, No, No, Yes, No, No, No, No, No, No, No, No, No, Yes, No…
$ Q3_12 <fct> No, Yes, No, No, No, No, No, Yes, Yes, No, Yes, No, Yes, No,…
$ Q3_13 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q3_14 <fct> No, No, No, No, Yes, No, Yes, Yes, No, No, No, No, No, No, N…
$ Q3_15 <fct> No, No, No, No, No, No, No, Yes, No, No, No, No, No, No, No,…
$ Q4 <fct> Field services, Ad hoc qual, Ad hoc qual, Data processing, T…
$ Q5 <fct> Yes, Yes, No, No, No, No, No, No, No, Yes, No, Yes, Yes, Yes…
$ Q6_1 <fct> Yes, Yes, No, No, No, No, No, No, No, Yes, No, Yes, Yes, Yes…
$ Q6_2 <fct> Yes, Yes, No, No, No, No, No, No, No, No, No, No, No, Yes, N…
$ Q6_3 <fct> Yes, Yes, No, No, No, No, No, No, No, Yes, No, No, No, Yes, …
$ Q6_4 <fct> No, Yes, No, No, No, No, No, No, No, Yes, No, No, No, No, No…
$ Q6_5 <fct> Yes, Yes, No, No, No, No, No, No, No, Yes, No, No, No, No, N…
$ Q6_6 <fct> No, No, No, No, No, No, No, No, No, Yes, No, No, No, No, No,…
$ Q6_7 <fct> No, Yes, No, No, No, No, No, No, No, Yes, No, No, No, No, No…
$ Q6_8 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q6_9 <fct> No, Yes, No, No, No, No, No, No, No, No, No, No, No, No, No,…
$ Q6_10 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q7 <fct> 40-60%, 20-40%, 20-40%, 40-60%, 80-100%, 0-20%, 0-20%, 0-20%…
$ Q8 <fct> Yes, Yes, No, Yes, Yes, No, Yes, No, Yes, Yes, Yes, Yes, Yes…
$ Q9_1 <fct> Occasionally, Never, Never, Never, Frequently, Never, Freque…
$ Q9_2 <fct> Frequently, Occasionally, Never, Occasionally, Frequently, N…
$ Q9_3 <fct> Never, Never, Never, Never, Occasionally, Never, Frequently,…
$ Q9_4 <fct> Never, Never, Never, Never, Never, Never, Never, Never, Neve…
$ Q9_5 <fct> Never, Never, Never, Never, Never, Never, Never, Never, Neve…
$ Q10 <fct> Yes, Yes, No, Yes, Yes, Yes, No, No, No, No, Yes, No, No, No…
$ Q11_1 <fct> Yes, Yes, No, Yes, Yes, No, Yes, Yes, No, No, Yes, Yes, No, …
$ Q11_2 <fct> No, No, No, Yes, No, Yes, Yes, No, No, No, Yes, No, Yes, No,…
$ Q11_3 <fct> No, No, No, Yes, No, No, No, No, No, No, No, No, No, No, No,…
$ Q11_4 <fct> No, No, No, Yes, No, Yes, No, No, No, No, Yes, No, Yes, No, …
$ Q11_5 <fct> No, No, Yes, No, No, No, No, No, Yes, Yes, No, No, No, Yes, …
$ Q11_other <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Q12_1 <fct> Very satisfied, Satisfied, Completely satisfied, Dissatisfie…
$ Q13_1 <fct> Satisfied, Very satisfied, Very satisfied, Satisfied, Neutra…
$ Q13_2 <fct> Very satisfied, Completely satisfied, Completely satisfied, …
$ Q13_3 <fct> Neutral, Very satisfied, Very satisfied, Neutral, Satisfied,…
$ Q13_4 <fct> Neutral, Satisfied, Neutral, Neutral, Neutral, Neutral, Neut…
$ Q14_1 <fct> Neutral, Satisfied, Neutral, Neutral, Satisfied, Neutral, Sa…
$ Q14_2 <fct> Very satisfied, Very satisfied, Very satisfied, Neutral, Ver…
$ Q14_3 <fct> Satisfied, Very satisfied, Very satisfied, Neutral, Neutral,…
$ Q14_4 <fct> Neutral, Neutral, Neutral, Dissatisfied, Dissatisfied, Neutr…
$ Q14_5 <fct> Very satisfied, Completely satisfied, Very satisfied, Neutra…
$ Q14_6 <fct> Very satisfied, Very satisfied, Neutral, Neutral, Satisfied,…
$ Q14_7 <fct> Satisfied, Neutral, Neutral, Neutral, Neutral, Neutral, Neut…
$ Q14_8 <fct> Satisfied, Very satisfied, Very satisfied, Neutral, Neutral,…
$ Q14_9 <fct> Satisfied, Very satisfied, Neutral, Neutral, Satisfied, Sati…
$ Q14_10 <fct> Satisfied, Very satisfied, Very satisfied, Neutral, Neutral,…
$ Q15_1 <fct> "Support via the e-group", "Support via the e-group", "Suppo…
$ Q15_2 <fct> "Opportunities for new business", "Events for networking, so…
$ Q15_3 <fct> "Opportunities for individual promotion", "Promotion of smal…
$ Q15_4 <fct> "Online resources ( website)", NA, "Promotion of smaller MR …
$ Q15_5 <fct> "Promotion of smaller MR consultant sector", NA, "Opportunit…
$ Q15_6 <fct> "Events for networking, socialising and learning", NA, "Oppo…
$ Q19_1 <fct> Rarely, Never, Never, Rarely, Never, Never, Never, Never, Ne…
$ Q19_2 <fct> Never, Occasionally, Rarely, Never, Never, Never, Never, Nev…
$ Q19_3 <fct> Never, Rarely, Never, Never, Never, Never, Occasionally, Nev…
$ Q19_4 <fct> Occasionally, Often / Always, Often / Always, Rarely, Occasi…
$ Q19_5 <fct> Often / Always, Occasionally, Occasionally, Rarely, Occasion…
$ Q19_6 <fct> Often / Always, Occasionally, Occasionally, Never, Occasiona…
$ Q20_1 <fct> Yes, No, No, Yes, No, No, No, Yes, No, No, Yes, No, No, No, …
$ Q20_2 <fct> No, No, No, Yes, No, Yes, No, No, No, No, Yes, No, No, No, Y…
$ Q20_3 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q20_4 <fct> No, No, No, No, No, Yes, No, No, No, No, Yes, Yes, No, No, N…
$ Q20_5 <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No, No, No,…
$ Q20_6 <fct> No, No, No, No, No, No, No, No, No, No, Yes, No, No, No, No,…
$ Q20_7 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q20_8 <fct> No, No, No, No, No, No, No, Yes, No, No, Yes, No, No, Yes, N…
$ Q20_9 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q20_10 <fct> No, No, No, Yes, No, No, No, No, No, No, No, No, No, No, No,…
$ Q20_other <fct> NA, NA, NA, NA, "I rarely attend business events in the even…
$ Q21_1 <fct> 2, 5 - Highly interested, 3 - Neutral / Not sure, 1 - Comple…
$ Q21_2 <fct> 4, 5 - Highly interested, 4, 1 - Completely uninterested, 2,…
$ Q21_3 <fct> 3 - Neutral / Not sure, 3 - Neutral / Not sure, 4, 1 - Compl…
$ Q21_4 <fct> 5 - Highly interested, 4, 1 - Completely uninterested, 1 - C…
$ Q21_5 <fct> 5 - Highly interested, 4, 1 - Completely uninterested, 1 - C…
$ Q21_6 <fct> 3 - Neutral / Not sure, 4, 3 - Neutral / Not sure, 3 - Neutr…
$ Q23_1 <fct> No, No, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes, No, Yes,…
$ Q23_2 <fct> Yes, Yes, No, No, No, No, No, No, No, No, No, No, No, No, Ye…
$ Q23_3 <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Q23_4 <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes, No, No,…
$ Q23_5 <fct> No, No, No, No, No, No, Yes, No, No, No, No, Yes, No, Yes, N…
$ Q23_other <fct> NA, "qrca", NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Q24 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Q25 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Q26_1 <ord> 21-49, 6-10, 6-10, 6-10, 21-49, 1-5, 6-10, NA, 6-10, 11-20, …
$ Q27_1 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Q27_2 <dbl> 1000, 700, 400, 300, NA, 450, 400, 200, 350, NA, 300, 350, 4…
$ Q30 <fct> Fieldwork, Consultancy, Consultancy, Data analysis, NA, Cons…
$ Q30_other <fct> NA, NA, NA, NA, Coach/Trainer, NA, NA, NA, NA, NA, NA, NA, N…
$ Q31 <fct> Sole trader, Limited company, Sole trader, Limited company, …
$ Q31_other <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Q32 <ord> Part-time (20-34 hours a week), Full-time (35 or more hours …
$ Q33 <fct> UK - Greater London, UK - Midlands / East of England / Wales…
$ Q35 <fct> Male, Female, Female, Female, Male, Male, Male, Female, Fema…
$ weight <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ size <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …Wrangling multiple columns
- Converting text to binary values
# A tibble: 215 × 109
id Q1_1 Q1_2 Q2 Q3_1 Q3_2 Q3_3 Q3_4 Q3_5 Q3_6 Q3_7 Q3_8 Q3_9
<dbl> <dbl> <dbl> <ord> <dbl> <fct> <fct> <fct> <fct> <fct> <fct> <fct> <fct>
1 3 8 2 2009 0 No No No No No No No No
2 5 35 12 Befo… 1 No No No No No No No Yes
3 6 34 12 Befo… 1 Yes No No No Yes No No No
4 11 20 9 2010 0 No No No No No No No No
5 13 20 3 2010 0 No No No No No No No No
6 15 36 20 Befo… 0 Yes No No No No No No Yes
7 21 12 2.5 2009 1 No No No No Yes Yes No No
8 22 11 0.5 2011 1 Yes Yes Yes Yes No No No No
9 23 18 3 2008 1 Yes Yes Yes Yes Yes No No Yes
10 25 24 8 2006 0 No No Yes Yes Yes No No Yes
# ℹ 205 more rows
# ℹ 96 more variables: Q3_10 <fct>, Q3_11 <fct>, Q3_12 <fct>, Q3_13 <fct>,
# Q3_14 <fct>, Q3_15 <fct>, Q4 <fct>, Q5 <fct>, Q6_1 <fct>, Q6_2 <fct>,
# Q6_3 <fct>, Q6_4 <fct>, Q6_5 <fct>, Q6_6 <fct>, Q6_7 <fct>, Q6_8 <fct>,
# Q6_9 <fct>, Q6_10 <fct>, Q7 <fct>, Q8 <fct>, Q9_1 <fct>, Q9_2 <fct>,
# Q9_3 <fct>, Q9_4 <fct>, Q9_5 <fct>, Q10 <fct>, Q11_1 <fct>, Q11_2 <fct>,
# Q11_3 <fct>, Q11_4 <fct>, Q11_5 <fct>, Q11_other <fct>, Q12_1 <fct>, …Wrangling multiple columns
- Converting all Q3_* columns
# A tibble: 215 × 109
id Q1_1 Q1_2 Q2 Q3_1 Q3_2 Q3_3 Q3_4 Q3_5 Q3_6 Q3_7 Q3_8 Q3_9
<dbl> <dbl> <dbl> <ord> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 3 8 2 2009 0 0 0 0 0 0 0 0 0
2 5 35 12 Befo… 1 0 0 0 0 0 0 0 1
3 6 34 12 Befo… 1 1 0 0 0 1 0 0 0
4 11 20 9 2010 0 0 0 0 0 0 0 0 0
5 13 20 3 2010 0 0 0 0 0 0 0 0 0
6 15 36 20 Befo… 0 1 0 0 0 0 0 0 1
7 21 12 2.5 2009 1 0 0 0 0 1 1 0 0
8 22 11 0.5 2011 1 1 1 1 1 0 0 0 0
9 23 18 3 2008 1 1 1 1 1 1 0 0 1
10 25 24 8 2006 0 0 0 1 1 1 0 0 1
# ℹ 205 more rows
# ℹ 96 more variables: Q3_10 <dbl>, Q3_11 <dbl>, Q3_12 <dbl>, Q3_13 <dbl>,
# Q3_14 <dbl>, Q3_15 <dbl>, Q4 <fct>, Q5 <fct>, Q6_1 <fct>, Q6_2 <fct>,
# Q6_3 <fct>, Q6_4 <fct>, Q6_5 <fct>, Q6_6 <fct>, Q6_7 <fct>, Q6_8 <fct>,
# Q6_9 <fct>, Q6_10 <fct>, Q7 <fct>, Q8 <fct>, Q9_1 <fct>, Q9_2 <fct>,
# Q9_3 <fct>, Q9_4 <fct>, Q9_5 <fct>, Q10 <fct>, Q11_1 <fct>, Q11_2 <fct>,
# Q11_3 <fct>, Q11_4 <fct>, Q11_5 <fct>, Q11_other <fct>, Q12_1 <fct>, …Wrangling multiple columns
- Q13_* columns are coded in Likert scale values in text
Q13_1 Q13_2 Q13_3
1 Satisfied Very satisfied Neutral
2 Very satisfied Completely satisfied Very satisfied
3 Very satisfied Completely satisfied Very satisfied
4 Satisfied Dissatisfied Neutral
5 Neutral Dissatisfied Satisfied
6 Satisfied Satisfied Neutral
7 Satisfied Very satisfied Completely satisfied
8 Satisfied Neutral Neutral
9 Very satisfied Completely satisfied Neutral
10 Satisfied Completely satisfied Satisfied
11 Very satisfied Satisfied Very satisfied
12 Satisfied Very satisfied Satisfied
13 Very satisfied Very satisfied Satisfied
14 Satisfied Satisfied Neutral
15 Satisfied Very satisfied Very satisfied
16 Satisfied Very satisfied Neutral
17 Satisfied Completely satisfied Very satisfied
18 Neutral Satisfied Neutral
19 Neutral Completely satisfied Very satisfied
20 Neutral Satisfied Neutral
21 Satisfied Completely satisfied Satisfied
22 Satisfied Satisfied Neutral
23 Satisfied Very satisfied Satisfied
24 Very satisfied Very satisfied Neutral
25 Neutral Satisfied Neutral
26 Satisfied Satisfied Neutral
27 Very satisfied Very satisfied Very satisfied
28 Satisfied Very satisfied Satisfied
29 Satisfied Very satisfied Dissatisfied
30 Satisfied Very satisfied Satisfied
31 Neutral Neutral Neutral
32 Satisfied Very satisfied Satisfied
33 Very satisfied Very satisfied Neutral
34 Very satisfied Very satisfied Very satisfied
35 Neutral Very satisfied Very satisfied
36 Very satisfied Completely satisfied Completely satisfied
37 Neutral Neutral Neutral
38 Very satisfied Completely satisfied Very satisfied
39 Satisfied Very satisfied Satisfied
40 Very satisfied Completely satisfied Very satisfied
41 Satisfied Very satisfied Satisfied
42 Neutral Very satisfied Neutral
43 Satisfied Very satisfied Satisfied
44 Very satisfied Completely satisfied Very satisfied
45 Very satisfied Very satisfied Neutral
46 Completely satisfied Completely satisfied Completely satisfied
47 Very satisfied Dissatisfied Neutral
48 Satisfied Satisfied Neutral
49 Satisfied Dissatisfied Very satisfied
50 Very satisfied Neutral Very satisfied
51 Neutral Satisfied Neutral
52 Satisfied Very satisfied Satisfied
53 Satisfied Very satisfied Satisfied
54 Neutral Completely satisfied Satisfied
55 Very satisfied Completely satisfied Very satisfied
56 Neutral Neutral Neutral
57 Completely satisfied Completely satisfied Very satisfied
58 Very satisfied Very satisfied Satisfied
59 Neutral Very satisfied Satisfied
60 Satisfied Satisfied Very satisfied
61 Satisfied Very satisfied Satisfied
62 Satisfied Very satisfied Satisfied
63 Neutral Dissatisfied Dissatisfied
64 Satisfied Very satisfied Neutral
65 Neutral Neutral Neutral
66 Completely satisfied Completely satisfied Completely satisfied
67 Very satisfied Completely satisfied Satisfied
68 Satisfied Very satisfied Satisfied
69 Neutral Completely satisfied Neutral
70 Satisfied Very satisfied Satisfied
71 Satisfied Satisfied Satisfied
72 Neutral Satisfied Neutral
73 Very satisfied Completely satisfied Neutral
74 Satisfied Satisfied Neutral
75 Dissatisfied Neutral Dissatisfied
76 Neutral Satisfied Satisfied
77 Neutral Very satisfied Satisfied
78 Neutral Completely satisfied Neutral
79 Satisfied Satisfied Satisfied
80 Neutral Satisfied Neutral
81 Very satisfied Very satisfied Very satisfied
82 Satisfied Completely satisfied Neutral
83 Very satisfied Completely satisfied Satisfied
84 Satisfied Very satisfied Very satisfied
85 Satisfied Completely satisfied Satisfied
86 Very satisfied Completely satisfied Satisfied
87 Satisfied Completely satisfied Satisfied
88 Neutral Satisfied Neutral
89 Satisfied Very satisfied Satisfied
90 Satisfied Very satisfied Satisfied
91 Very satisfied Very satisfied Very satisfied
92 Neutral Very satisfied Neutral
93 Satisfied Completely satisfied Very satisfied
94 Satisfied Satisfied Neutral
95 Neutral Very satisfied Neutral
96 Very satisfied Satisfied Neutral
97 Satisfied Satisfied Satisfied
98 Satisfied Very satisfied Satisfied
99 Neutral Satisfied Neutral
100 Neutral Completely satisfied Neutral
101 Very satisfied Completely satisfied Very satisfied
102 Satisfied Completely satisfied Neutral
103 Very satisfied Satisfied Very satisfied
104 Neutral Very satisfied Neutral
105 Satisfied Very satisfied Satisfied
106 Satisfied Very satisfied Satisfied
107 Neutral Very satisfied Neutral
108 Satisfied Completely satisfied Neutral
109 Neutral Very satisfied Neutral
110 Very satisfied Completely satisfied Very satisfied
111 Satisfied Very satisfied Very satisfied
112 Satisfied Satisfied Satisfied
113 Neutral Very satisfied Satisfied
114 Completely satisfied Very satisfied Neutral
115 Neutral Very satisfied Neutral
116 Satisfied Completely satisfied Satisfied
117 Satisfied Very satisfied Satisfied
118 Dissatisfied Very satisfied Neutral
119 Neutral Satisfied Neutral
120 Neutral Neutral Neutral
121 Satisfied Very satisfied Satisfied
122 Satisfied Very satisfied Very dissatisfied
123 Neutral Completely satisfied Neutral
124 Neutral Very satisfied Satisfied
125 Neutral Very satisfied Satisfied
126 Very satisfied Very satisfied Very satisfied
127 Satisfied Completely satisfied Neutral
128 Neutral Very satisfied Very satisfied
129 Neutral Neutral Neutral
130 Satisfied Very satisfied Satisfied
131 Satisfied Very satisfied Satisfied
132 Satisfied Very satisfied Satisfied
133 Satisfied Satisfied Neutral
134 Very satisfied Completely satisfied Very satisfied
135 Dissatisfied Satisfied Very dissatisfied
136 Very satisfied Completely satisfied Satisfied
137 Satisfied Very satisfied Neutral
138 Completely satisfied Completely satisfied Very satisfied
139 Neutral Neutral Neutral
140 Satisfied Very satisfied Neutral
141 Dissatisfied Satisfied Neutral
142 Completely satisfied Completely satisfied Very satisfied
143 Neutral Neutral Satisfied
144 Neutral Neutral Neutral
145 Satisfied Satisfied Neutral
146 Satisfied Completely satisfied Very satisfied
147 Satisfied Very satisfied Neutral
148 Dissatisfied Very satisfied Neutral
149 Very satisfied Very satisfied Very satisfied
150 Satisfied Neutral Satisfied
151 Satisfied Satisfied Neutral
152 Satisfied Satisfied Neutral
153 Satisfied Very satisfied Neutral
154 Very dissatisfied Satisfied Neutral
155 Very satisfied Satisfied Neutral
156 Dissatisfied Satisfied Neutral
157 Neutral Satisfied Neutral
158 Very satisfied Dissatisfied Neutral
159 Satisfied Very satisfied Satisfied
160 Neutral Completely satisfied Neutral
161 Satisfied Very satisfied Neutral
162 Satisfied Completely satisfied Neutral
163 Neutral Satisfied Very satisfied
164 Satisfied Satisfied Neutral
165 Satisfied Very satisfied Neutral
166 Very satisfied Very satisfied Very satisfied
167 Neutral Very satisfied Neutral
168 Very satisfied Satisfied Satisfied
169 Very satisfied Completely satisfied Very satisfied
170 Satisfied Satisfied Satisfied
171 Dissatisfied Satisfied Neutral
172 Satisfied Very satisfied Neutral
173 Very satisfied Very satisfied Very satisfied
174 Neutral Satisfied Neutral
175 Satisfied Satisfied Neutral
176 Satisfied Completely satisfied Satisfied
177 Satisfied Satisfied Very satisfied
178 Very satisfied Very satisfied Satisfied
179 Very satisfied Very satisfied Satisfied
180 Very satisfied Very satisfied Neutral
181 Neutral Very satisfied Satisfied
182 Completely satisfied Completely satisfied Completely satisfied
183 Neutral Satisfied Neutral
184 Satisfied Very satisfied Satisfied
185 Satisfied Very satisfied Satisfied
186 Satisfied Very satisfied Satisfied
187 Very satisfied Very satisfied Very satisfied
188 Satisfied Very satisfied Neutral
189 Neutral Very satisfied Neutral
190 Satisfied Satisfied Neutral
191 Dissatisfied Very satisfied Neutral
192 Satisfied Completely satisfied Satisfied
193 Satisfied Very satisfied Satisfied
194 Very satisfied Very satisfied Very satisfied
195 Satisfied Very satisfied Satisfied
196 Satisfied Dissatisfied Satisfied
197 Very satisfied Very satisfied Very satisfied
198 Neutral Very satisfied Neutral
199 Satisfied Very satisfied Satisfied
200 Very satisfied Dissatisfied Very satisfied
201 Satisfied Satisfied Satisfied
202 Completely satisfied Completely satisfied Completely satisfied
203 Satisfied Satisfied Neutral
204 Dissatisfied Satisfied Neutral
205 Satisfied Very satisfied Neutral
206 Very satisfied Very satisfied Satisfied
207 Neutral Satisfied Neutral
208 Neutral Dissatisfied Neutral
209 Very satisfied Very satisfied Very satisfied
210 Very satisfied Very satisfied Very satisfied
211 Neutral Satisfied Neutral
212 Satisfied Satisfied Satisfied
213 Satisfied Satisfied Satisfied
214 Satisfied Satisfied Satisfied
215 Neutral Very satisfied Neutral
Q13_4
1 Neutral
2 Satisfied
3 Neutral
4 Neutral
5 Neutral
6 Neutral
7 Neutral
8 Neutral
9 Completely satisfied
10 Satisfied
11 Satisfied
12 Neutral
13 Satisfied
14 Neutral
15 Neutral
16 Very dissatisfied
17 Very satisfied
18 Neutral
19 Completely satisfied
20 Satisfied
21 Satisfied
22 Neutral
23 Dissatisfied
24 Neutral
25 Neutral
26 Neutral
27 Neutral
28 Satisfied
29 Satisfied
30 Very satisfied
31 Neutral
32 Dissatisfied
33 Satisfied
34 Very satisfied
35 Very satisfied
36 Satisfied
37 Neutral
38 Satisfied
39 Neutral
40 Very satisfied
41 Satisfied
42 Neutral
43 Neutral
44 Very satisfied
45 Neutral
46 Completely satisfied
47 Neutral
48 Neutral
49 Neutral
50 Neutral
51 Neutral
52 Neutral
53 Neutral
54 Satisfied
55 Neutral
56 Neutral
57 Very satisfied
58 Satisfied
59 Neutral
60 Very satisfied
61 Neutral
62 Satisfied
63 Satisfied
64 Neutral
65 Neutral
66 Satisfied
67 Very satisfied
68 Satisfied
69 Satisfied
70 Neutral
71 Neutral
72 Neutral
73 Very satisfied
74 Satisfied
75 Satisfied
76 Neutral
77 Neutral
78 Dissatisfied
79 Satisfied
80 Neutral
81 Very satisfied
82 Neutral
83 Very satisfied
84 Very satisfied
85 Neutral
86 Neutral
87 Very satisfied
88 Dissatisfied
89 Neutral
90 Very satisfied
91 Neutral
92 Neutral
93 Very satisfied
94 Neutral
95 Neutral
96 Neutral
97 Neutral
98 Neutral
99 Neutral
100 Satisfied
101 Satisfied
102 Dissatisfied
103 Dissatisfied
104 Satisfied
105 Satisfied
106 Satisfied
107 Satisfied
108 Neutral
109 Satisfied
110 Very satisfied
111 Satisfied
112 Satisfied
113 Satisfied
114 Neutral
115 Neutral
116 Neutral
117 Satisfied
118 Neutral
119 Satisfied
120 Neutral
121 Satisfied
122 Very satisfied
123 Neutral
124 Neutral
125 Satisfied
126 Satisfied
127 Satisfied
128 Dissatisfied
129 Neutral
130 Satisfied
131 Neutral
132 Satisfied
133 Very satisfied
134 Satisfied
135 Satisfied
136 Neutral
137 Neutral
138 Neutral
139 Satisfied
140 Neutral
141 Neutral
142 Very satisfied
143 Satisfied
144 Neutral
145 Neutral
146 Satisfied
147 Neutral
148 Dissatisfied
149 Very satisfied
150 Neutral
151 Neutral
152 Dissatisfied
153 Neutral
154 Dissatisfied
155 Neutral
156 Neutral
157 Satisfied
158 Neutral
159 Satisfied
160 Satisfied
161 Neutral
162 Satisfied
163 Very satisfied
164 Neutral
165 Dissatisfied
166 Very satisfied
167 Neutral
168 Satisfied
169 Satisfied
170 Satisfied
171 Neutral
172 Neutral
173 Very satisfied
174 Neutral
175 Satisfied
176 Satisfied
177 Satisfied
178 Very satisfied
179 Satisfied
180 Satisfied
181 Neutral
182 Satisfied
183 Neutral
184 Satisfied
185 Very satisfied
186 Neutral
187 Very satisfied
188 Neutral
189 Neutral
190 Neutral
191 Satisfied
192 Neutral
193 Neutral
194 Neutral
195 Neutral
196 Satisfied
197 Neutral
198 Neutral
199 Satisfied
200 Satisfied
201 Satisfied
202 Satisfied
203 Neutral
204 Neutral
205 Satisfied
206 Neutral
207 Satisfied
208 Neutral
209 Very satisfied
210 Very satisfied
211 Neutral
212 Neutral
213 Satisfied
214 Satisfied
215 DissatisfiedWrangling multiple columns
- Manually recode all columns
membersurvey %>%
select(starts_with("Q13_")) %>%
mutate(
Q13_1 = case_when(
Q13_1 == "Completely dissatisfied" ~ -3,
Q13_1 == "Very dissatisfied" ~ -2,
Q13_1 == "Dissatisfied" ~ -1,
Q13_1 == "Neutral" ~ 0,
Q13_1 == "Satisfied" ~ 1,
Q13_1 == "Very satisfied" ~ 2,
Q13_1 == "Completely satisfied" ~ 3
),
Q13_2 = case_when(
Q13_2 == "Completely dissatisfied" ~ -3,
Q13_2 == "Very dissatisfied" ~ -2,
Q13_2 == "Dissatisfied" ~ -1,
Q13_2 == "Neutral" ~ 0,
Q13_2 == "Satisfied" ~ 1,
Q13_2 == "Very satisfied" ~ 2,
Q13_2 == "Completely satisfied" ~ 3
),
Q13_3 = case_when(
Q13_3 == "Completely dissatisfied" ~ -3,
Q13_3 == "Very dissatisfied" ~ -2,
Q13_3 == "Dissatisfied" ~ -1,
Q13_3 == "Neutral" ~ 0,
Q13_3 == "Satisfied" ~ 1,
Q13_3 == "Very satisfied" ~ 2,
Q13_3 == "Completely satisfied" ~ 3
),
Q13_4 = case_when(
Q13_4 == "Completely dissatisfied" ~ -3,
Q13_4 == "Very dissatisfied" ~ -2,
Q13_4 == "Dissatisfied" ~ -1,
Q13_4 == "Neutral" ~ 0,
Q13_4 == "Satisfied" ~ 1,
Q13_4 == "Very satisfied" ~ 2,
Q13_4 == "Completely satisfied" ~ 3
)
)- Output
Q13_1 Q13_2 Q13_3 Q13_4
1 1 2 0 0
2 2 3 2 1
3 2 3 2 0
4 1 -1 0 0
5 0 -1 1 0
6 1 1 0 0
7 1 2 3 0
8 1 0 0 0
9 2 3 0 3
10 1 3 1 1
11 2 1 2 1
12 1 2 1 0
13 2 2 1 1
14 1 1 0 0
15 1 2 2 0
16 1 2 0 -2
17 1 3 2 2
18 0 1 0 0
19 0 3 2 3
20 0 1 0 1
21 1 3 1 1
22 1 1 0 0
23 1 2 1 -1
24 2 2 0 0
25 0 1 0 0
26 1 1 0 0
27 2 2 2 0
28 1 2 1 1
29 1 2 -1 1
30 1 2 1 2
31 0 0 0 0
32 1 2 1 -1
33 2 2 0 1
34 2 2 2 2
35 0 2 2 2
36 2 3 3 1
37 0 0 0 0
38 2 3 2 1
39 1 2 1 0
40 2 3 2 2
41 1 2 1 1
42 0 2 0 0
43 1 2 1 0
44 2 3 2 2
45 2 2 0 0
46 3 3 3 3
47 2 -1 0 0
48 1 1 0 0
49 1 -1 2 0
50 2 0 2 0
51 0 1 0 0
52 1 2 1 0
53 1 2 1 0
54 0 3 1 1
55 2 3 2 0
56 0 0 0 0
57 3 3 2 2
58 2 2 1 1
59 0 2 1 0
60 1 1 2 2
61 1 2 1 0
62 1 2 1 1
63 0 -1 -1 1
64 1 2 0 0
65 0 0 0 0
66 3 3 3 1
67 2 3 1 2
68 1 2 1 1
69 0 3 0 1
70 1 2 1 0
71 1 1 1 0
72 0 1 0 0
73 2 3 0 2
74 1 1 0 1
75 -1 0 -1 1
76 0 1 1 0
77 0 2 1 0
78 0 3 0 -1
79 1 1 1 1
80 0 1 0 0
81 2 2 2 2
82 1 3 0 0
83 2 3 1 2
84 1 2 2 2
85 1 3 1 0
86 2 3 1 0
87 1 3 1 2
88 0 1 0 -1
89 1 2 1 0
90 1 2 1 2
91 2 2 2 0
92 0 2 0 0
93 1 3 2 2
94 1 1 0 0
95 0 2 0 0
96 2 1 0 0
97 1 1 1 0
98 1 2 1 0
99 0 1 0 0
100 0 3 0 1
101 2 3 2 1
102 1 3 0 -1
103 2 1 2 -1
104 0 2 0 1
105 1 2 1 1
106 1 2 1 1
107 0 2 0 1
108 1 3 0 0
109 0 2 0 1
110 2 3 2 2
111 1 2 2 1
112 1 1 1 1
113 0 2 1 1
114 3 2 0 0
115 0 2 0 0
116 1 3 1 0
117 1 2 1 1
118 -1 2 0 0
119 0 1 0 1
120 0 0 0 0
121 1 2 1 1
122 1 2 -2 2
123 0 3 0 0
124 0 2 1 0
125 0 2 1 1
126 2 2 2 1
127 1 3 0 1
128 0 2 2 -1
129 0 0 0 0
130 1 2 1 1
131 1 2 1 0
132 1 2 1 1
133 1 1 0 2
134 2 3 2 1
135 -1 1 -2 1
136 2 3 1 0
137 1 2 0 0
138 3 3 2 0
139 0 0 0 1
140 1 2 0 0
141 -1 1 0 0
142 3 3 2 2
143 0 0 1 1
144 0 0 0 0
145 1 1 0 0
146 1 3 2 1
147 1 2 0 0
148 -1 2 0 -1
149 2 2 2 2
150 1 0 1 0
151 1 1 0 0
152 1 1 0 -1
153 1 2 0 0
154 -2 1 0 -1
155 2 1 0 0
156 -1 1 0 0
157 0 1 0 1
158 2 -1 0 0
159 1 2 1 1
160 0 3 0 1
161 1 2 0 0
162 1 3 0 1
163 0 1 2 2
164 1 1 0 0
165 1 2 0 -1
166 2 2 2 2
167 0 2 0 0
168 2 1 1 1
169 2 3 2 1
170 1 1 1 1
171 -1 1 0 0
172 1 2 0 0
173 2 2 2 2
174 0 1 0 0
175 1 1 0 1
176 1 3 1 1
177 1 1 2 1
178 2 2 1 2
179 2 2 1 1
180 2 2 0 1
181 0 2 1 0
182 3 3 3 1
183 0 1 0 0
184 1 2 1 1
185 1 2 1 2
186 1 2 1 0
187 2 2 2 2
188 1 2 0 0
189 0 2 0 0
190 1 1 0 0
191 -1 2 0 1
192 1 3 1 0
193 1 2 1 0
194 2 2 2 0
195 1 2 1 0
196 1 -1 1 1
197 2 2 2 0
198 0 2 0 0
199 1 2 1 1
200 2 -1 2 1
201 1 1 1 1
202 3 3 3 1
203 1 1 0 0
204 -1 1 0 0
205 1 2 0 1
206 2 2 1 0
207 0 1 0 1
208 0 -1 0 0
209 2 2 2 2
210 2 2 2 2
211 0 1 0 0
212 1 1 1 0
213 1 1 1 1
214 1 1 1 1
215 0 2 0 -1Wrangling multiple columns
- Functional approach: write a simple function!
Wrangling multiple columns
- Functional approach: apply the function across columns
membersurvey %>%
mutate(across(starts_with("Q13_"), convert_text_to_num)) %>%
select(starts_with("Q13_")) Q13_1 Q13_2 Q13_3 Q13_4
1 1 2 0 0
2 2 3 2 1
3 2 3 2 0
4 1 -1 0 0
5 0 -1 1 0
6 1 1 0 0
7 1 2 3 0
8 1 0 0 0
9 2 3 0 3
10 1 3 1 1
11 2 1 2 1
12 1 2 1 0
13 2 2 1 1
14 1 1 0 0
15 1 2 2 0
16 1 2 0 -2
17 1 3 2 2
18 0 1 0 0
19 0 3 2 3
20 0 1 0 1
21 1 3 1 1
22 1 1 0 0
23 1 2 1 -1
24 2 2 0 0
25 0 1 0 0
26 1 1 0 0
27 2 2 2 0
28 1 2 1 1
29 1 2 -1 1
30 1 2 1 2
31 0 0 0 0
32 1 2 1 -1
33 2 2 0 1
34 2 2 2 2
35 0 2 2 2
36 2 3 3 1
37 0 0 0 0
38 2 3 2 1
39 1 2 1 0
40 2 3 2 2
41 1 2 1 1
42 0 2 0 0
43 1 2 1 0
44 2 3 2 2
45 2 2 0 0
46 3 3 3 3
47 2 -1 0 0
48 1 1 0 0
49 1 -1 2 0
50 2 0 2 0
51 0 1 0 0
52 1 2 1 0
53 1 2 1 0
54 0 3 1 1
55 2 3 2 0
56 0 0 0 0
57 3 3 2 2
58 2 2 1 1
59 0 2 1 0
60 1 1 2 2
61 1 2 1 0
62 1 2 1 1
63 0 -1 -1 1
64 1 2 0 0
65 0 0 0 0
66 3 3 3 1
67 2 3 1 2
68 1 2 1 1
69 0 3 0 1
70 1 2 1 0
71 1 1 1 0
72 0 1 0 0
73 2 3 0 2
74 1 1 0 1
75 -1 0 -1 1
76 0 1 1 0
77 0 2 1 0
78 0 3 0 -1
79 1 1 1 1
80 0 1 0 0
81 2 2 2 2
82 1 3 0 0
83 2 3 1 2
84 1 2 2 2
85 1 3 1 0
86 2 3 1 0
87 1 3 1 2
88 0 1 0 -1
89 1 2 1 0
90 1 2 1 2
91 2 2 2 0
92 0 2 0 0
93 1 3 2 2
94 1 1 0 0
95 0 2 0 0
96 2 1 0 0
97 1 1 1 0
98 1 2 1 0
99 0 1 0 0
100 0 3 0 1
101 2 3 2 1
102 1 3 0 -1
103 2 1 2 -1
104 0 2 0 1
105 1 2 1 1
106 1 2 1 1
107 0 2 0 1
108 1 3 0 0
109 0 2 0 1
110 2 3 2 2
111 1 2 2 1
112 1 1 1 1
113 0 2 1 1
114 3 2 0 0
115 0 2 0 0
116 1 3 1 0
117 1 2 1 1
118 -1 2 0 0
119 0 1 0 1
120 0 0 0 0
121 1 2 1 1
122 1 2 -2 2
123 0 3 0 0
124 0 2 1 0
125 0 2 1 1
126 2 2 2 1
127 1 3 0 1
128 0 2 2 -1
129 0 0 0 0
130 1 2 1 1
131 1 2 1 0
132 1 2 1 1
133 1 1 0 2
134 2 3 2 1
135 -1 1 -2 1
136 2 3 1 0
137 1 2 0 0
138 3 3 2 0
139 0 0 0 1
140 1 2 0 0
141 -1 1 0 0
142 3 3 2 2
143 0 0 1 1
144 0 0 0 0
145 1 1 0 0
146 1 3 2 1
147 1 2 0 0
148 -1 2 0 -1
149 2 2 2 2
150 1 0 1 0
151 1 1 0 0
152 1 1 0 -1
153 1 2 0 0
154 -2 1 0 -1
155 2 1 0 0
156 -1 1 0 0
157 0 1 0 1
158 2 -1 0 0
159 1 2 1 1
160 0 3 0 1
161 1 2 0 0
162 1 3 0 1
163 0 1 2 2
164 1 1 0 0
165 1 2 0 -1
166 2 2 2 2
167 0 2 0 0
168 2 1 1 1
169 2 3 2 1
170 1 1 1 1
171 -1 1 0 0
172 1 2 0 0
173 2 2 2 2
174 0 1 0 0
175 1 1 0 1
176 1 3 1 1
177 1 1 2 1
178 2 2 1 2
179 2 2 1 1
180 2 2 0 1
181 0 2 1 0
182 3 3 3 1
183 0 1 0 0
184 1 2 1 1
185 1 2 1 2
186 1 2 1 0
187 2 2 2 2
188 1 2 0 0
189 0 2 0 0
190 1 1 0 0
191 -1 2 0 1
192 1 3 1 0
193 1 2 1 0
194 2 2 2 0
195 1 2 1 0
196 1 -1 1 1
197 2 2 2 0
198 0 2 0 0
199 1 2 1 1
200 2 -1 2 1
201 1 1 1 1
202 3 3 3 1
203 1 1 0 0
204 -1 1 0 0
205 1 2 0 1
206 2 2 1 0
207 0 1 0 1
208 0 -1 0 0
209 2 2 2 2
210 2 2 2 2
211 0 1 0 0
212 1 1 1 0
213 1 1 1 1
214 1 1 1 1
215 0 2 0 -1Wrangling multiple columns
across()also works withsummarize()!
Short break
Any question so far?
A segue to tibble
Tibble is a extremely flexible data structure
- A column can be numeric, character, logical, date…etc
A segue to tibble
Tibble is a extremely flexible data structure
- It can also be a list of vectors
A segue to tibble
Tibble is a extremely flexible data structure
- Or even a list of tibbles (or df, or anything)!
A segue to tibble
Tibble is a extremely flexible data structure
We usually think of tidy data this way:
Columns = variables (some characteristics of obs.)
Rows = observations (e.g., people, countries)
Another way to exploit tibble
Columns = different inputs/arguments to a function
Rows = different specifications/values to those inputs
Then
map()a function over those specifications
Mapping a function in a tibble
- A realistic example: running regressions
# A tibble: 10 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Uganda Africa 1957 42.6 6675501 774.
2 Korea, Rep. Asia 1982 67.1 39326000 5623.
3 Algeria Africa 1972 54.5 14760787 4183.
4 Panama Americas 1987 71.5 2253639 7035.
5 Iceland Europe 1997 79.0 271192 28061.
6 Saudi Arabia Asia 1952 39.9 4005677 6460.
7 Puerto Rico Americas 1987 74.6 3444468 12281.
8 Slovenia Europe 1982 71.1 1861252 17867.
9 Zambia Africa 1967 47.8 3900000 1777.
10 Yemen, Rep. Asia 1992 55.6 13367997 1879.Mapping a function in a tibble
- A realistic example: running regressions
Call:
lm(formula = m1, data = gapminder)
Residuals:
Min 1Q Median 3Q Max
-82.754 -7.758 2.176 8.225 18.426
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.396e+01 3.150e-01 171.29 <2e-16 ***
gdpPercap 7.649e-04 2.579e-05 29.66 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 10.49 on 1702 degrees of freedom
Multiple R-squared: 0.3407, Adjusted R-squared: 0.3403
F-statistic: 879.6 on 1 and 1702 DF, p-value: < 2.2e-16Mapping a function in a tibble
- A realistic example: running regressions
Mapping a function in a tibble
- I can run a regression inside a tibble
Mapping a function in a tibble
- I can run a regression inside a tibble
Mapping a function in a tibble
- I can run a regression inside a tibble
Mapping a function in a tibble
- I can run a regression inside a tibble
lm_tbl <-
tibble(
data = list(gapminder),
lm_res = map(data, ~ lm(lifeExp ~ gdpPercap, data = .x)),
lm_res_tidy = map(lm_res, ~ tidy(.x, conf.int = TRUE))
)
lm_tbl$lm_res_tidy[[1]]
# A tibble: 2 × 7
term estimate std.error statistic p.value conf.low conf.high
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 54.0 0.315 171. 0 53.3 54.6
2 gdpPercap 0.000765 0.0000258 29.7 3.57e-156 0.000714 0.000815Mapping a function in a tibble
Why? – We can run many regressions simultaneously and wrangle all output efficiently
Mapping a function in a tibble
- Running multiple regressions
Mapping a function in a tibble
- Running multiple regressions
gapminder %>%
group_by(continent) %>%
nest() %>%
mutate(lm_res = map(data, ~ lm(lifeExp ~ gdpPercap, data = .x)))# A tibble: 5 × 3
# Groups: continent [5]
continent data lm_res
<fct> <list> <list>
1 Asia <tibble [396 × 5]> <lm>
2 Europe <tibble [360 × 5]> <lm>
3 Africa <tibble [624 × 5]> <lm>
4 Americas <tibble [300 × 5]> <lm>
5 Oceania <tibble [24 × 5]> <lm> Mapping a function in a tibble
- Running multiple regressions
gapminder %>%
group_by(continent) %>%
nest() %>%
mutate(lm_res = map(data, ~ lm(lifeExp ~ gdpPercap, data = .x)),
lm_res_tidy = map(lm_res, ~ tidy(.x, conf.int = TRUE)))# A tibble: 5 × 4
# Groups: continent [5]
continent data lm_res lm_res_tidy
<fct> <list> <list> <list>
1 Asia <tibble [396 × 5]> <lm> <tibble [2 × 7]>
2 Europe <tibble [360 × 5]> <lm> <tibble [2 × 7]>
3 Africa <tibble [624 × 5]> <lm> <tibble [2 × 7]>
4 Americas <tibble [300 × 5]> <lm> <tibble [2 × 7]>
5 Oceania <tibble [24 × 5]> <lm> <tibble [2 × 7]>Mapping a function in a tibble
- Running multiple regressions
Mapping a function in a tibble
- Running multiple regressions
# A tibble: 10 × 8
# Groups: continent [5]
continent term estimate std.error statistic p.value conf.low conf.high
<fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Asia (Interce… 5.75e+1 0.633 90.8 2.45e-266 5.63e+1 58.8
2 Asia gdpPercap 3.23e-4 0.0000393 8.21 3.29e- 15 2.45e-4 0.000400
3 Europe (Interce… 6.53e+1 0.330 198. 0 6.47e+1 66.0
4 Europe gdpPercap 4.53e-4 0.0000192 23.6 4.05e- 75 4.16e-4 0.000491
5 Africa (Interce… 4.58e+1 0.420 109. 0 4.50e+1 46.7
6 Africa gdpPercap 1.38e-3 0.000117 11.7 7.60e- 29 1.15e-3 0.00161
7 Americas (Interce… 5.88e+1 0.672 87.5 1.33e-214 5.75e+1 60.2
8 Americas gdpPercap 8.16e-4 0.0000702 11.6 5.45e- 26 6.78e-4 0.000954
9 Oceania (Interce… 6.37e+1 0.729 87.4 1.87e- 29 6.22e+1 65.2
10 Oceania gdpPercap 5.71e-4 0.0000371 15.4 2.99e- 13 4.94e-4 0.000648Mapping a function in a tibble
- Running multiple regressions
Mapping a function in a tibble
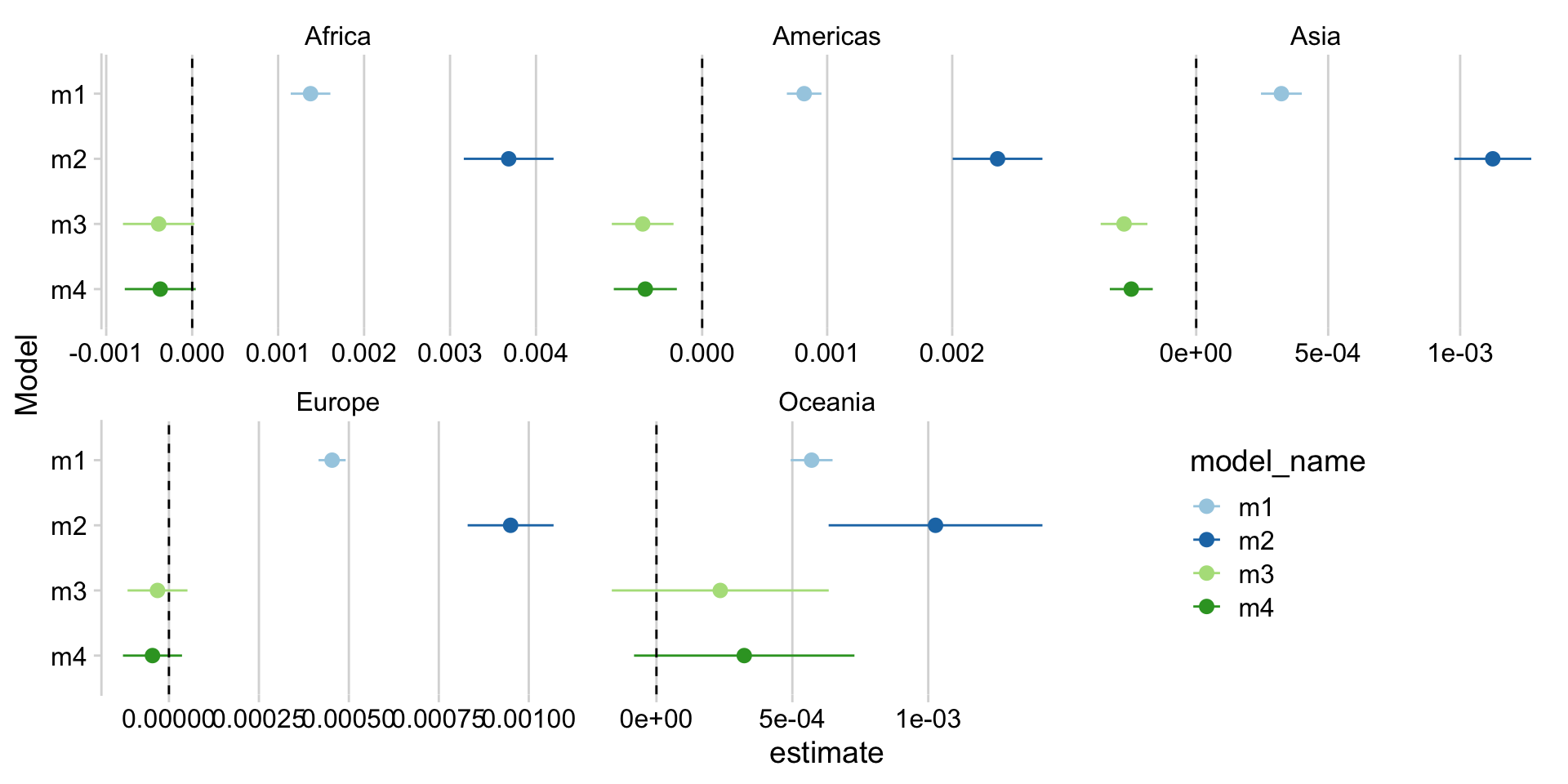
- Visualizing multiple regressions
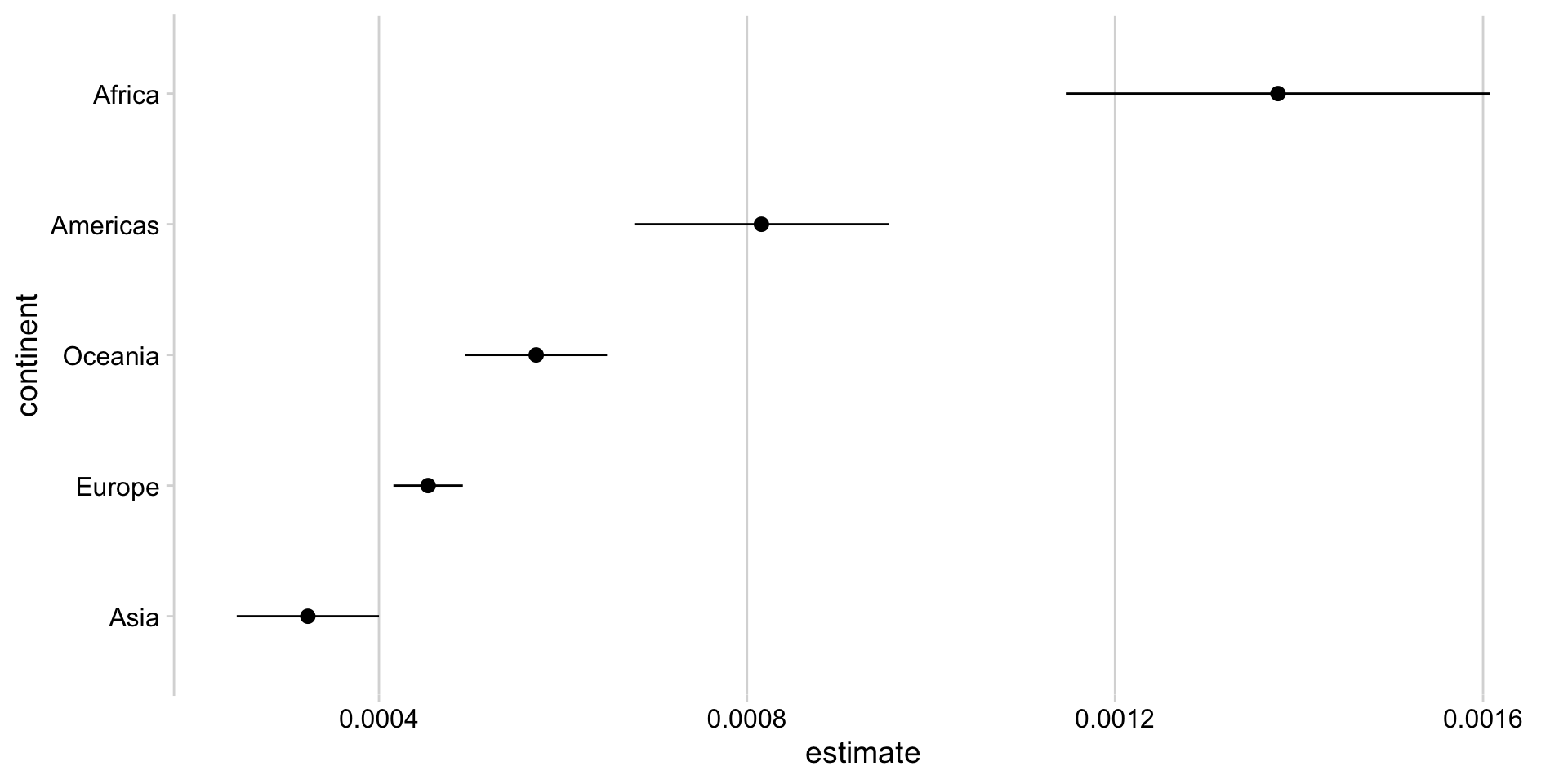
Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
m1 <- lifeExp ~ gdpPercap
m2 <- lifeExp ~ gdpPercap + I(gdpPercap^2)
m3 <- lifeExp ~ gdpPercap + log(gdpPercap)
m4 <- lifeExp ~ gdpPercap + log(gdpPercap) + log(pop)
model_list <- list("m1" = m1, "m2" = m2, "m3" = m3, "m4" = m4)
model_list$m1
lifeExp ~ gdpPercap
$m2
lifeExp ~ gdpPercap + I(gdpPercap^2)
$m3
lifeExp ~ gdpPercap + log(gdpPercap)
$m4
lifeExp ~ gdpPercap + log(gdpPercap) + log(pop)Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
# A tibble: 5 × 3
# Groups: continent [5]
continent data model
<fct> <list> <list>
1 Asia <tibble [396 × 5]> <named list [4]>
2 Europe <tibble [360 × 5]> <named list [4]>
3 Africa <tibble [624 × 5]> <named list [4]>
4 Americas <tibble [300 × 5]> <named list [4]>
5 Oceania <tibble [24 × 5]> <named list [4]>Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
gapminder %>%
group_by(continent) %>% nest() %>%
mutate(model = list(model_list)) %>%
unnest_longer(model, values_to = "formula", indices_to = "model_name")# A tibble: 20 × 4
# Groups: continent [5]
continent data formula model_name
<fct> <list> <named list> <chr>
1 Asia <tibble [396 × 5]> <formula> m1
2 Asia <tibble [396 × 5]> <formula> m2
3 Asia <tibble [396 × 5]> <formula> m3
4 Asia <tibble [396 × 5]> <formula> m4
5 Europe <tibble [360 × 5]> <formula> m1
6 Europe <tibble [360 × 5]> <formula> m2
7 Europe <tibble [360 × 5]> <formula> m3
8 Europe <tibble [360 × 5]> <formula> m4
9 Africa <tibble [624 × 5]> <formula> m1
10 Africa <tibble [624 × 5]> <formula> m2
11 Africa <tibble [624 × 5]> <formula> m3
12 Africa <tibble [624 × 5]> <formula> m4
13 Americas <tibble [300 × 5]> <formula> m1
14 Americas <tibble [300 × 5]> <formula> m2
15 Americas <tibble [300 × 5]> <formula> m3
16 Americas <tibble [300 × 5]> <formula> m4
17 Oceania <tibble [24 × 5]> <formula> m1
18 Oceania <tibble [24 × 5]> <formula> m2
19 Oceania <tibble [24 × 5]> <formula> m3
20 Oceania <tibble [24 × 5]> <formula> m4 Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
gapminder %>%
group_by(continent) %>% nest() %>%
mutate(model = list(model_list)) %>%
unnest_longer(model, values_to = "formula", indices_to = "model_name") %>%
mutate(lm_res = map2(formula, data, lm)) # A tibble: 20 × 5
# Groups: continent [5]
continent data formula model_name lm_res
<fct> <list> <named list> <chr> <named list>
1 Asia <tibble [396 × 5]> <formula> m1 <lm>
2 Asia <tibble [396 × 5]> <formula> m2 <lm>
3 Asia <tibble [396 × 5]> <formula> m3 <lm>
4 Asia <tibble [396 × 5]> <formula> m4 <lm>
5 Europe <tibble [360 × 5]> <formula> m1 <lm>
6 Europe <tibble [360 × 5]> <formula> m2 <lm>
7 Europe <tibble [360 × 5]> <formula> m3 <lm>
8 Europe <tibble [360 × 5]> <formula> m4 <lm>
9 Africa <tibble [624 × 5]> <formula> m1 <lm>
10 Africa <tibble [624 × 5]> <formula> m2 <lm>
11 Africa <tibble [624 × 5]> <formula> m3 <lm>
12 Africa <tibble [624 × 5]> <formula> m4 <lm>
13 Americas <tibble [300 × 5]> <formula> m1 <lm>
14 Americas <tibble [300 × 5]> <formula> m2 <lm>
15 Americas <tibble [300 × 5]> <formula> m3 <lm>
16 Americas <tibble [300 × 5]> <formula> m4 <lm>
17 Oceania <tibble [24 × 5]> <formula> m1 <lm>
18 Oceania <tibble [24 × 5]> <formula> m2 <lm>
19 Oceania <tibble [24 × 5]> <formula> m3 <lm>
20 Oceania <tibble [24 × 5]> <formula> m4 <lm> Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
gapminder %>%
group_by(continent) %>% nest() %>%
mutate(model = list(model_list)) %>%
unnest_longer(model, values_to = "formula", indices_to = "model_name") %>%
mutate(lm_res = map2(formula, data, lm),
lm_res = map(lm_res, ~ tidy(.x, conf.int = TRUE))) # A tibble: 20 × 5
# Groups: continent [5]
continent data formula model_name lm_res
<fct> <list> <named list> <chr> <named list>
1 Asia <tibble [396 × 5]> <formula> m1 <tibble [2 × 7]>
2 Asia <tibble [396 × 5]> <formula> m2 <tibble [3 × 7]>
3 Asia <tibble [396 × 5]> <formula> m3 <tibble [3 × 7]>
4 Asia <tibble [396 × 5]> <formula> m4 <tibble [4 × 7]>
5 Europe <tibble [360 × 5]> <formula> m1 <tibble [2 × 7]>
6 Europe <tibble [360 × 5]> <formula> m2 <tibble [3 × 7]>
7 Europe <tibble [360 × 5]> <formula> m3 <tibble [3 × 7]>
8 Europe <tibble [360 × 5]> <formula> m4 <tibble [4 × 7]>
9 Africa <tibble [624 × 5]> <formula> m1 <tibble [2 × 7]>
10 Africa <tibble [624 × 5]> <formula> m2 <tibble [3 × 7]>
11 Africa <tibble [624 × 5]> <formula> m3 <tibble [3 × 7]>
12 Africa <tibble [624 × 5]> <formula> m4 <tibble [4 × 7]>
13 Americas <tibble [300 × 5]> <formula> m1 <tibble [2 × 7]>
14 Americas <tibble [300 × 5]> <formula> m2 <tibble [3 × 7]>
15 Americas <tibble [300 × 5]> <formula> m3 <tibble [3 × 7]>
16 Americas <tibble [300 × 5]> <formula> m4 <tibble [4 × 7]>
17 Oceania <tibble [24 × 5]> <formula> m1 <tibble [2 × 7]>
18 Oceania <tibble [24 × 5]> <formula> m2 <tibble [3 × 7]>
19 Oceania <tibble [24 × 5]> <formula> m3 <tibble [3 × 7]>
20 Oceania <tibble [24 × 5]> <formula> m4 <tibble [4 × 7]>Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
# A tibble: 60 × 9
# Groups: continent [5]
continent model_name term estimate std.error statistic p.value conf.low
<fct> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Asia m1 (Interc… 5.75e+1 6.33e- 1 90.8 2.45e-266 5.63e+1
2 Asia m1 gdpPerc… 3.23e-4 3.93e- 5 8.21 3.29e- 15 2.45e-4
3 Asia m2 (Interc… 5.38e+1 6.20e- 1 86.8 1.65e-258 5.26e+1
4 Asia m2 gdpPerc… 1.12e-3 7.42e- 5 15.1 4.05e- 41 9.78e-4
5 Asia m2 I(gdpPe… -1.03e-8 8.47e-10 -12.1 7.01e- 29 -1.19e-8
6 Asia m3 (Interc… -6.16e+0 3.67e+ 0 -1.68 9.38e- 2 -1.34e+1
7 Asia m3 gdpPerc… -2.73e-4 4.50e- 5 -6.07 3.04e- 9 -3.62e-4
8 Asia m3 log(gdp… 8.47e+0 4.84e- 1 17.5 3.55e- 51 7.52e+0
9 Asia m4 (Interc… -4.56e+1 5.63e+ 0 -8.10 6.83e- 15 -5.67e+1
10 Asia m4 gdpPerc… -2.46e-4 4.14e- 5 -5.94 6.19e- 9 -3.27e-4
# ℹ 50 more rows
# ℹ 1 more variable: conf.high <dbl>Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
many_models_tbl %>%
select(continent, model_name, lm_res) %>%
unnest(lm_res) %>%
filter(term == "gdpPercap")# A tibble: 20 × 9
# Groups: continent [5]
continent model_name term estimate std.error statistic p.value conf.low
<fct> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Asia m1 gdpPercap 3.23e-4 0.0000393 8.21 3.29e-15 2.45e-4
2 Asia m2 gdpPercap 1.12e-3 0.0000742 15.1 4.05e-41 9.78e-4
3 Asia m3 gdpPercap -2.73e-4 0.0000450 -6.07 3.04e- 9 -3.62e-4
4 Asia m4 gdpPercap -2.46e-4 0.0000414 -5.94 6.19e- 9 -3.27e-4
5 Europe m1 gdpPercap 4.53e-4 0.0000192 23.6 4.05e-75 4.16e-4
6 Europe m2 gdpPercap 9.50e-4 0.0000607 15.6 2.28e-42 8.30e-4
7 Europe m3 gdpPercap -3.17e-5 0.0000423 -0.750 4.54e- 1 -1.15e-4
8 Europe m4 gdpPercap -4.57e-5 0.0000417 -1.10 2.74e- 1 -1.28e-4
9 Africa m1 gdpPercap 1.38e-3 0.000117 11.7 7.60e-29 1.15e-3
10 Africa m2 gdpPercap 3.68e-3 0.000266 13.8 3.31e-38 3.16e-3
11 Africa m3 gdpPercap -3.90e-4 0.000212 -1.84 6.60e- 2 -8.05e-4
12 Africa m4 gdpPercap -3.71e-4 0.000210 -1.77 7.76e- 2 -7.84e-4
13 Americas m1 gdpPercap 8.16e-4 0.0000702 11.6 5.45e-26 6.78e-4
14 Americas m2 gdpPercap 2.36e-3 0.000182 13.0 9.81e-31 2.00e-3
15 Americas m3 gdpPercap -4.76e-4 0.000125 -3.79 1.79e- 4 -7.23e-4
16 Americas m4 gdpPercap -4.55e-4 0.000128 -3.55 4.50e- 4 -7.07e-4
17 Oceania m1 gdpPercap 5.71e-4 0.0000371 15.4 2.99e-13 4.94e-4
18 Oceania m2 gdpPercap 1.03e-3 0.000189 5.43 2.19e- 5 6.33e-4
19 Oceania m3 gdpPercap 2.35e-4 0.000192 1.22 2.35e- 1 -1.64e-4
20 Oceania m4 gdpPercap 3.23e-4 0.000194 1.66 1.12e- 1 -8.25e-5
# ℹ 1 more variable: conf.high <dbl>Mapping a function in a tibble
- Many specifications: 4 models x 5 continents
many_models_tbl %>%
select(continent, model_name, lm_res) %>%
unnest(lm_res) %>%
filter(term == "gdpPercap") %>%
ggplot(aes(y = fct_rev(model_name),
color = model_name,
x = estimate, xmin = conf.low, xmax = conf.high)) +
geom_pointrange() +
geom_vline(xintercept = 0, linetype = 2) +
facet_wrap(~ continent, scales = "free_x") +
labs(y = "Model") +
scale_color_brewer(type = "qual", palette = 3) +
cowplot::theme_minimal_vgrid() +
theme(legend.position = c(0.75,0.25))Mapping a function in a tibble
- Many specifications: 4 models x 5 continents